A Comprehensive Survey of Mamba Architectures for Medical Image Analysis: Classification, Segmentation, Restoration and Beyond
Nice Survey paper on Mamba. Mamba revolutionizes medical image analysis with efficient long-range dependency modeling and versatile applications.
Nice Survey paper on Mamba.
Mamba revolutionizes medical image analysis with efficient long-range dependency modeling and versatile applications.
Mamba is a specialized case of State Space Models (SSMs) that has emerged as an alternative to transformer-based architectures for medical image analysis tasks.
Key differences include:
Mamba has linear time complexity compared to the quadratic complexity of transformers, allowing it to process longer sequences more efficiently.
Mamba can handle long-range dependencies without attention mechanisms, enabling faster inference with less memory usage.
Mamba shows strong performance in merging multimodal data, which is beneficial for medical applications.
Compared to CNNs, Mamba is better at capturing long-range dependencies while maintaining efficiency.
The main types of Mamba architectures for medical image analysis include:
Pure Mamba:
Vision Mamba (ViM): Uses bidirectional SSM for image processing.
VMamba: Incorporates Visual State Space (VSS) block with 2D selective scanning.
Plain Mamba: Non-hierarchical SSM similar to Vision Transformers.
U-Net Variants:
Mamba-UNet: Integrates Mamba blocks into U-Net architecture.
VM-UNet: Uses Visual Mamba blocks in encoder and decoder.
H-vmunet: Incorporates higher-order Visual State Space modules.
Hybrid Architectures:
Combines Mamba with other techniques:
Convolutions: e.g., HC-Mamba, nnMamba
Attention/Transformers: e.g., Weak-Mamba-UNet
Recurrence: e.g., VMRNN
Graph Neural Networks: e.g., for whole slide image analysis
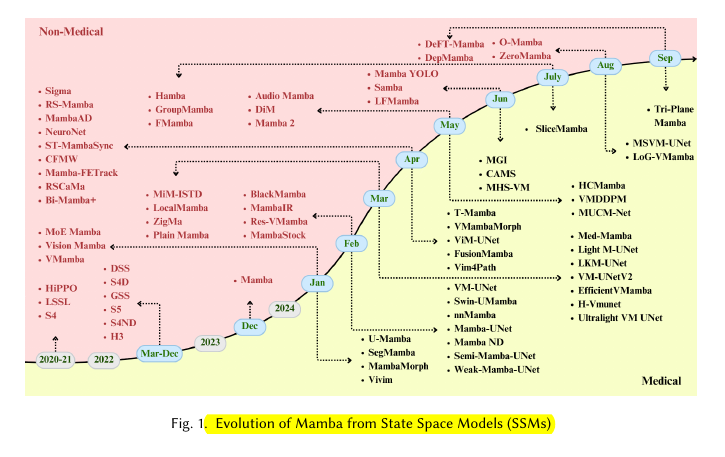
The evolution from S4 to Mamba involved several key steps:
S4: Introduced efficient computation methods for SSMs and used structured matrices for parameter A.
S5: Simplified S4, introduced learnable timescale parameter, and used Zero-Order Hold discretization.
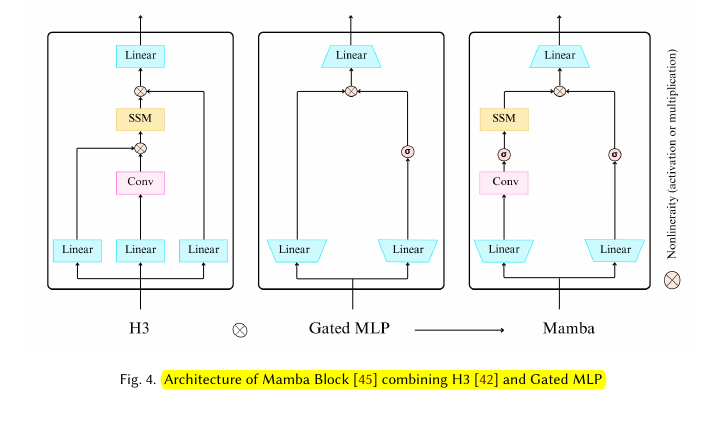
S6 (Mamba): Incorporated selective scanning mechanism, hardware-aware algorithm, and improved initialization.
Mamba architectures differ in how they integrate Mamba with existing neural network designs, balance local and global feature extraction, and adapt to specific medical imaging tasks.
The core concepts behind State Space Models (SSMs)?
State Space Models (SSMs) are a class of models that convert 1D input sequences into N-dimensional continuous latent states, which are then projected back to 1D outputs. The core concepts include:
Continuous-time formulation: SSMs are defined by differential equations in continuous time.
Discretization: For practical use with discrete inputs, SSMs are discretized using methods like Zero-Order Hold.
Key parameters: SSMs are defined by parameters A, B, C, D which control state transitions and input/output projections.